----
In a previous post about viewing functional data from the Human Connectome Project (HCP) I described downloading the images then viewing them with the Connectome Workbench. This post describes a way to extract the timecourses for specific surface vertices using code, rather than one-at-a-time in the Workbench.
First, working with surfaces means working with GIFTI and CIFTI files. Many developers are creating functions that can read these files, but software is less mature and harder to find than for reading NIfTI files (MATLAB, FSL and NiBabel seem furthest along; I couldn't find one for R). The GIFTI library for MATLAB worked great ... except with HCP-derived files.Guillaume Flandin very kindly (and quickly!) changed his code to work with HCP files, making it ignore the spec-inconsistent part of those headers. By the time you read this, the files might be fixed, but for now (19 May 2014), if the GIFTI library for MATLAB gives you errors with HCP files (but not others),
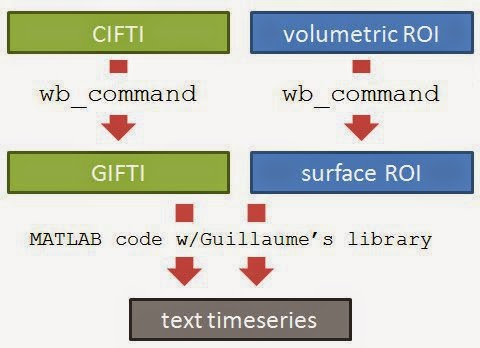
Here is an overview of the process. The logic is the simple: figure out which voxels/vertices are needed, then get the timecourses for those voxels/vertices. For HCP functional surface images there are two preliminary steps: extracting the part of the brain we want from the CIFTI file (green), and finding which vertices we want (such as by constructing a surface ROI mask, blue).
Note that it's not necessary to create a ROI to extract the timecourses; if you know which vertices you need by some other means you can read them directly in MATLAB once the GIFTI has been loaded.
unpack the CIFTI
It works best for me to think of HCP CIFTI files as archives containing the functional data for the whole brain, as a mixture of surfaces (for the cortical sheet) and volumes (for sub-cortical structures). Before reading the functional data we need to unpack the CIFTI, making GIFTI or NIfTI files, as appropriate for the part of the brain we want; the wb_command -cifti-separate program does this "unpacking" (see this post for notes about downloading HCP data, and this one about working with the Workbench wb_command command-line program). For example, if we want the functional time courses for a ROI on the left hemisphere surface, we run this at the command line:wb_command -cifti-separate in.dtseries.nii COLUMN -metric CORTEX_LEFT out.func.giiwhere in.dtseries.nii is the path and filename of an HCP tfMRI CIFTI file (e.g. tfMRI_MOTOR_LR_Atlas.dtseries.nii), and out.func.gii is the path and filename of the GIFTI we want the program to create. The command's help has more options and explanation; in brief CORTEX_LEFT indicates which anatomical structure to unpack, COLUMN gets timecourses, and -metric is because CORTEX_LEFT is stored as a surface.
identify the desired vertices
There is not a one-to-one correspondence between volumetric voxels and surface vertices in the HCP functional datasetwb_command -volume-to-surface-mapping roi.nii.gz atlas.surf.gii roi.func.gii -enclosingwhere roi.nii.gz is the volumetric ROI mask, atlas.surf.gii is an atlas surface for the hemisphere containing the ROI, and roi.func.gii is the GIFTI metric file that will be created. This only works if the surface atlas, volumetric ROI, and functional CIFTI are all aligned to the same space. For the HCP data, the MNINonLinear images for each person are aligned to the MNI atlas, specifically matching the conte69 32k atlas. Thus, I made the volumetric ROI on an MNI template brain, then used Conte69.L.midthickness.32k_fs_LR.surf.gii for the atlas.surf.gii in the -volume-to-surface-mapping call. So long as the headers are correct in roi.nii.gz (i.e. the voxel size, origin, etc are correct) the ROI should be in the correct place in roi.func.gii, but view it in Workbench to be sure.
extract the vertex timecourses
Finally, we can read both the ROI and functional data GIFTI files into MATLAB, reading the vertex indices from the ROI then saving the timecourses as text:addpath 'C:/Program Files/MATLAB/gifti-1.4'; % path to GIFTI library
roi = gifti(['d:/temp/roi.func.gii']); % load the ROI GIFTI
inds = find(roi.cdata > 0); % find is like which: get the vertex indices
wm = gifti([inpath 'out.func.gii']); % load the functional GIFTI
tmp = wm.cdata(inds,:); % get those indices' timcourses
csvwrite(['d:/temp/out.csv'], tmp); % write as a csv text file
Now, out.csv has one column for each timepoint and one row for each vertex in the ROI.
confirming the match
We can check that the values match by opening out.func.gii in both Workbench and MATLAB:In the image I clicked on vertex 5336 in Workbench (red arrow), which is the vertex at position 5337 in the MATLAB .cdata array (purple arrow), since MATLAB is one-based but Workbench is zero-based. The first two values of the data series shown in the Workbench Information window (red underline) match those shown in MATLAB (purple underline).
Note: I could only get Workbench to show the first two values of the timeseries if I set the little blue-arrowed button to 2; otherwise it would display only the first value. Clicking through the little blue-arrow box changes the display to different timepoints, but doesn't change the Information window that I could tell (I'm using Workbench 0.85). Note also that the "charting" interface has changed from my previous post; now you need to go through the "Chart" radio button on the upper left of the View window (right under the first tab name on my screen); I couldn't get it to write out the full timeseries in Workbench itself.
UPDATE (20 May 2014): Tim Coalson suggested that by convention the output files should be named roi.func.gii and out.func.gii, not roi.shape.gii and out.dtseries.gii as I originally wrote; I changed the commands accordingly. Tim also pointed me to the program wb_command -surface-closest-vertex, which will return the closest vertex to an arbitrary 3d coordinate. He suggests that to go the other way (from a vertex in a .surf.gii to 3d coordinates) you "could look at the coordinate of a particular vertex, and back-convert through the nifti sform to get the real-valued voxel "indices" it resides at (real-valued because it could be a third of a voxel to the right of a voxel center, etc)."



