 Back in April 2013 I posted instructions for how to plot a NIfTI image in caret. However, caret isn't under active development any more: it has been superseded by/transformed into the Connectome Workbench.
Back in April 2013 I posted instructions for how to plot a NIfTI image in caret. However, caret isn't under active development any more: it has been superseded by/transformed into the Connectome Workbench.As always, please let me know (or comment) with corrections, or if you know of easier ways to accomplish anything I describe here. I performed these steps today (19 September 2013), using workbench_windows64.beta_0.83, and have updated as needed for later versions.
downloading
The installation procedure for the Workbench is quite similar to that for caret. First, download the appropriate version, then unzip it to wherever you want to run it from. Like caret, you don't "install" Workbench but just unzip the download then double-click the executable. On my windows box I unzipped it into d:/, so double-clickThe program doesn't come with a tutorial or manual, but a tutorial pdf and dataset are linked from the same page as the program itself. I strongly suggest you go through at least the first chapter of the tutorial, since it steps through the main parts of the software's interface, giving pretty clear click-by-click directions and explanations.
getting a blank brain
While the tutorial clearly explains how to navigate through and manipulate the sample dataset, it doesn't explain how to get a blank brain and overlay a NIfTI image, which is what I want to do to duplicate what I did with caret. So here goes.This test NIfTI image is aligned to MNI space. My caret-saavy coworker suggested that the Conte69 atlas is an appropriate underlay for this image. Follow the links from this page to download the Workbench-compatible version of the atlas (32k_fs_LR mesh). Like installing Workbench itself, the atlas is downloaded as a zip, which you should unpack into a sensible directory; I put mine into a subdirectory of the main workbench directory.
Look in the atlas directory: there will be a single .spec file (Conte69_atlas-v2.LR.32k_fs_LR.wb.spec). Make a copy of this file, and rename the copy to something describing the anatomy + overlay we're making, like test.spec. This way, we can save the overlay and anatomy together (and so avoid doing the imports again) without overwriting the original files.
Start the Workbench. In Windows, I double-click
The first window closes and a new one appears, listing all of the files linked in the spec file. For now, let's just load them all, so don't change anything, and click the Load button. My computer gives a little dialog showing the progress as the files are loaded (along with a warning that something is missing), which disappears and the main Workbench appears, with a blank brain.
changing the main viewing window
Here's what the blank brain looks like on my computer. (NOTE 2 June 2016: The program has a few more buttons in Workbench version 1.2.0, but the features I describe are all still present. Hopefully someday I'll investigate all the new features!) Workbench is quite similar to what we saw in caret, but more hemispheres and a lot fewer clicks and menus to get to this point.I'm not going to go into detail about manipulating the view, since I think chapter 1 of the tutorial explains it pretty well.
Briefly, the parts I marked in orange at right are the main controls for adjusting the display: the dropboxes in the "Montage Selection" area control how inflated the anatomy is, and the checkboxes which hemispheres are shown. Like in caret, you can use the mouse to rotate the images (and zoom), or click the little buttons in the "View" and "Orientation" panes to switch to standard views.
The bottom section (marked with orange arrows) controls viewing the overlays. The conte69 atlas set includes some overlays, listed in the File dropboxes, and shown (or not) by the little On checkboxes. Click around (and read the tutorial) to figure out what these do.
overlaying a NIfTI image: preparation
Now that we have the blank brain we can put our clusters on it. In my caret directions I describe a series of menu choices, starting with clicking on Attributes, then Map Volume(s) to Surface(s) in the top menus. But, these menus are not present in the Workbench, so we have to use the command line. What we will do is use the command line program to turn our NIfTI into two .shape.gii files, which we can then open, view, and add to the test.spec file in the Workbench GUI.So, to run a command line program we need a command line. I don't like messing with paths, so I navigated to the directory containing wb_command.exe (on my machine, /bin_windows64/, same as workbench.exe). We're going to use the -volume-to-surface-mapping function; typing wb_command -volume-to-surface-mapping at the command prompt should cause it to print out the help file and details about how the mapping is done. As its help explains, the -volume-to-surface-mapping function needs the NIfTI we're converting (fakeBrain.nii.gz), what we want the output to be called, and what surface we're mapping to.
In this case, we want to put the NIfTI onto the conte69 blank brain (which we loaded into Workbench above). The -volume-to-surface-mapping function uses a surf.gii file to specify the surface; several of these came in the conte69 atlas set. I want a modest level of inflation, so I decide on the "midthickness" surf.gii files (there are also very inflated ones, etc.). Note that there are two midthickness surf.gii files: one for the left hemisphere, one for the right. Thus, we will run the -volume-to-surface-mapping function twice on the NIfTI, making one shape.gii file for the left hemisphere and another for the right.
Now we're ready, so run these two lines at the command prompt (change the paths, obviously):
wb_command -volume-to-surface-mapping d:/temp/fakeBrain.nii.gz d:/temp/Conte69_atlas-v2.LR.32k_fs_LR.wb/32k_ConteAtlas_v2/Conte69.L.midthickness.32k_fs_LR.surf.gii d:/temp/fakeBrainL.shape.gii -trilinear
wb_command -volume-to-surface-mapping d:/temp/fakeBrain.nii.gz d:/temp/Conte69_atlas-v2.LR.32k_fs_LR.wb/32k_ConteAtlas_v2/Conte69.R.midthickness.32k_fs_LR.surf.gii d:/temp/fakeBrainR.shape.gii -trilinear
If all goes well, there won't be any output at the command line, but two files will be created: fakeBrainL.shape.gii and fakeBrainR.shape.gii.
actually overlaying the NIfTI image
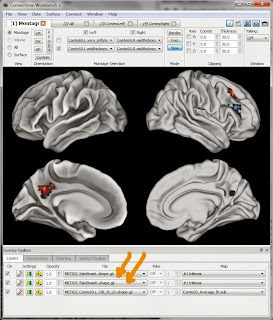 Now, we can actually overlay our NIfTI image (well, the shape.gii files created from the .nii.gz). In the Workbench program's top menus click on File then Open File. Change the dropdown box at the bottom of the screen so that it's looking for Files of type "Metric Files (*.func.gii *.shape.gii)". Navigate and select both fakeBrainL.shape.gii and fakeBrainR.shape.gii.
Now, we can actually overlay our NIfTI image (well, the shape.gii files created from the .nii.gz). In the Workbench program's top menus click on File then Open File. Change the dropdown box at the bottom of the screen so that it's looking for Files of type "Metric Files (*.func.gii *.shape.gii)". Navigate and select both fakeBrainL.shape.gii and fakeBrainR.shape.gii.The Workbench screen won't visibly change, but don't fear! There should now be two new entries in the File dropboxes at the bottom of the screen (marked here with big orange arrows); select them, click the corresponding On checkbox, and the blobs should appear, as shown here. The little boxes to the left of the dropboxes (Settings and Opacity) adjust colors, show scales, and more rows, etc.
saving the changes to test.spec
If you want, you can add the shape.gii files to the .spec file. Then, instead of having to open them each time you can open the single test.spec file. Click "File" then "Save/Manage Files ..." from the top menus. A window will appear listing everything in the test.spec, as well as the two Metric files we added (fakeBrainL.shape.gii and fakeBrainR.shape.gii). The little checkboxes in the "In Spec" column will not be checked for these files, because we just opened them. Check the boxes for these files in both the "Save" and "In Spec" columns, as well as the very top checkbox in the "Save" column for the spec file itself, and then click the "Save Checked Files" button at the bottom of the dialog. The checks in the Save column (and red warning lettering) will disappear, but those in the "In Spec file" column will not, showing that they were saved into test.spec. Click "Close" at the bottom of the dialog.Select File, then Exit from the top menu. A warning should appear that you are exiting; if one appears that there are unsaved changes, check that everything was marked properly in the "Save/Manage Files ..." dialog. Open up the Workbench again. test.spec should now be in the list of "Recent Spec Files"; opening it will bring up the program with fakeBrainL.shape.gii and fakeBrainR.shape.gii already in the Overlay File dropboxes, without having to load them separately.
comments
While having to go to the command line is a bit of a hassle, after a bit of exploration I've found the Workbench a bit easier to use than caret to do this plotting. The Connectome Workbench is under very active development, so I'm sure some of these directions will soon be obsolete. But for now, give it a try!NOTICE (8 February 2018): The material in this post is outdated. If you're trying to learn Workbench you should consult this new tutorial for a general introduction, and this one for instructions about creating a surface from a volumetric image.
UPDATE 2 August 2017: added a reference to the new getting-started tutorial.
UPDATE 2 June 2016: Made a few light edits and confirmed that these instructions work with Workbench version 1.2.0.
UPDATE 9 February 2014: The same procedure will work to plot a NIfTI that has integer values (e.g. a ROI mask), but the resulting surface image will have strange shading around the edges of the ROIs. To avoid this, use -enclosing in the command to wb_command instead of -trilinear.

Hi JO,
ReplyDeleteThanks for the tutorial!
I have one question for your: Conte69 brain does not contain the cerebellum. Do you know an alternative to Conte69 that I can use for overlaying my nifri file?
Glad you found the tutorials helpful!
DeleteI suspect you were the one who posted this question to the connectome workbench list, but if not, it was answered here: http://www.mail-archive.com/hcp-users@humanconnectome.org/msg00846.html
Dear Joe,
ReplyDeleteThank you so much for your very helpful tutorial. I have one question that I hope you can help me with.
When I project a statistical map as an overlay to the Conte69 Atlas of workbench, a big part of it is not projected on the surface. By checking the Vol/Surf Outline on the overlay toolbox, it seems that the borders of the surface do not include all the grey matter. Therefore any activation outside the borders (in the grey matter) are projected on the volumes but not projected on the surface. Is there any solution to that ?
Thank you.
Glad the tutorial helped!
DeleteThe conte69 surface should capture the grey matter very precisely, but the projection from volume to surface when the NIfTI is just a functional volume is sometimes not very precise at all.
I suggest you start by checking the image you're projecting onto the surface: is it normalized to the MNI atlas (or another atlas, in which case you need a different surface)? Is all of the header information (voxel size, orientation, etc) correct ( wb_command -file-information might be helpful here)?
If all looks correct with your volume, you could try changing some of the wb_command -volume-to-surface-mapping options (http://humanconnectome.org/documentation/workbench-command/command-volume-to-surface-mapping.html); changing the ribbon or mapping method might help.
If none of this works, you could try the hcp-users mailing list; I think there have been some similar threads. Good luck, and please reply again letting us know what works! :)
Dear Jozet, thank you for your tutorial!
ReplyDeleteI was wondering, if there is a way to combine the separate hemisphere files into one? The reason I wonder is that when I visualize some statistics, I often want to show 5% strongest voxels, but when the threshold adjustment needs to be done on each hemisphere separately, I just can't get the same colorbar and same min-max values for it. Maybe you know some other way to do it?
Thank you in advance for any advice :)
I haven't been able to play with the new version of Workbench yet (they're up to version 1.1.1 now). But in version 1.0 I didn't ever combine the hemispheres; I always worked with separate files. But I was able to get matching scales for each hemisphere by changing around the values in the Overlay and Map Settings box (click the little wrench icons at the bottom of the screen). For example, to show values between 0.6 and 1, with the colorbar from 0.6 to 1 even if no voxels have a value of 1, select "Fixed" in the bottom left of the dialog, then set the Pos Min to 0.6 and Pos Max to 1. Then, put a Threshold on with a High of 0.6, and Show Data Outside Thresholds.
DeleteThis would be easier with a screenshot; when I get a tutorial with the new version of Workbench up I'll try adding a description of this. Good luck!
Ah, this worked perfectly! Thank you!
DeleteDear Jo,
ReplyDeleteI would like to do functional and structural connectivity analysis and i was using the pipelines suggested by https://github.com/Washington-University/Pipelines, for preprocessing DTI and fcMRI data, could you please let me know how should i approach functional connectivity problem on workbench, i have used SPM for modelling but now i am learning to use workbench , and i find it very difficult to start.
Thanks
Sorry, but I've never tried any connectivity analyses, in or out of Workbench, and don't know of any connectivity-specific tutorials to suggest. Perhaps working though my tutorials will help you get familiar with workbench, at least. Good luck!
DeleteHi Jo,
ReplyDeleteThanks for the tutorial! It is exactly what I was looking for. However, the Conte69 atlas is no longer available in the link you attached (the sumsDB is down). Do you happen to have a copy of it that you can send over, or maybe you know how to reach it within the hcp?
Thank you!
Tamar
The sumsDB worked fine for me when I tested it just now. For alternatives, googling Conte69_atlas-v2.LR.32k_fs_LR.wb brought up https://github.com/mgxd/brain_plots/tree/master/32k_ConteAtlas_v2, which looks to have the same files.
Delete