My go-to program for separating clusters out of an image is the Clusterize routine in AFNI. This little tutorial steps you through getting a NIfTI image into AFNI, using Clusterize, then getting a NIfTI out again. A word of warning: be sure to check laterality in the post-Clusterize NIfTI; sometimes things get flipped when you use multiple analysis programs. Also, I have a Windows box, so run AFNI within NeuroDebian (you should, too, especially if you run Windows), as the screenshots and notes below reflect.
First, you need to get your NIfTI image into AFNI. Since I use NeuroDebian I started by putting the NIfTI I want to open into the for_afni subdirectory of the shared folder. Then you need to tell AFNI which directory to find the images in, which you do by clicking the Read button in the DataDir window (top red arrow). The Read Session window appears (right side of the screenshot, and, since I'm in NeuroDebian, I find my for_afni subdirectory under /home/brain/host/. Clicking the Set button (bottom red arrow) makes AFNI look for images in the directory.
Now we need to display the image that we want to clusterize. The image needs to be loaded as an OverLay, but AFNI is happiest if it has both an UnderLay and the OverLay, which are loaded via the circled buttons. Clicking the UnderLay button brings up a list of images, from both the for_afni subdirectory (since it was Read in the previous step) and standard anatomies (in my installation). In the screenshot I picked a standard anatomy; it also works if you use the overlay for the underlay (but you need something for the underlay). Then click the OverLay button and select the image you want to cluster. After setting both images you should see colored blobs on top of a greyscale background image: the colored image (the OverLay) will be the one clustered. Then click the Define OverLay button (arrow) to bring up the display shown in the upper right corner of the screenshot.
Next, set the threshold so that only the voxels you want to cluster are shown. Here, my overlay image consists of integers, and I want to identify clusters of at least 10 voxels with the value of 6 or higher. The screenshot shows how to set the threshold of 6: I put the little ** dropdown menu to 1 so that the values in the color slider are the actual numerical values (rather than a statistic). Next, I uncheck the autoRange box, also since I want the slider to be the actual numerical values. Finally, I move the slider (top arrow) to be exactly at 6 (use the up and down arrows for fine-tuning). The overlay changed as I moved the slider: now only voxels with values of 6 or larger are shown, and the overlay color scaling shifted.
Now we can do the clustering. First, click the Clear button (circled), in case any previous clustering is still in memory. Then click the Clusterize button (also circled), which brings up the menu dialog box (shown at left). Adjust the NN level and Voxels boxes to match your clustering parameters; the screenshot is set to find clusters with at least 10 voxels, and voxels must share a side to be in the same cluster. Click the Set button to close the menu dialog box. The main display won't change, except that the Rpt button (circled) will be enabled.
Clicking the Rpt button brings up the AFNI Cluster Results dialog box, as shown here. The display shows that AFNI found 8 clusters in my mask, ranging in size from 277 to 10 voxels. The coordinates are shown for the peak voxel in each cluster (since the XYZ dropdown is set to Peak), and clicking the Jump button in each row changes the coordinates in the display accordingly. To save the clustered version of the image, type a name into the box to the left of the SaveMsk button (marked with an arrow), then click the SaveMsk button. It doesn't look like anything happened, but there should now be a pair of images in the AFNI output directory (/brain/ by default in NeuroDebian) starting with the name specified.
Last, we need to convert the clustered mask back in NIfTI. I do this at the command line with 3dcopy. Not liking to mess about with configuration files, when I first open up the terminal I run . /etc/afni/afni.sh so that it can find 3dcopy. In the screenshot the terminal window opened up in the /brain/ directory, which is where the AFNI files are, so running 3dcopy outImage_mask+tlrc outImage.nii.gz writes the NIfTI file in /brain/ as well. Then I copy-paste outImage.nii.gz into /host/for_afni/ so that I can get to the file in Windows.
None of these steps are particularly difficult, but navigating back and forth can be a bit tricky, and the steps need to be done in the proper order. Good luck!
UPDATE 19 November 2015: You can also use afni at the command prompt for clustering; try the different options of 3dmerge and 3dmask_tool.
Wednesday, September 24, 2014
Thursday, September 18, 2014
demo: R code to perform a voxelwise t-test
It's easy to perform a voxelwise t-test (t-test at each voxel individually). Programs for mass-univariate analysis (like SPM and fsl) of course do this (and much more), but sometimes you just want to do a simple t-test across subjects at each voxel.
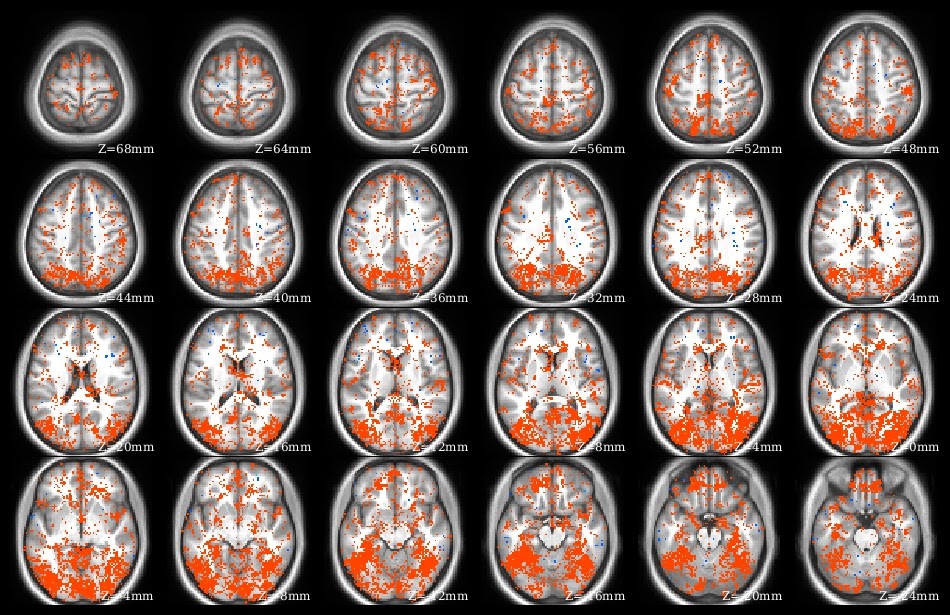 The demo code linked in this post does a voxelwise t-test in R. It takes as input a set of 3d NIfTI files, where each NIfTI is assumed to come from a different person, each voxel of which contains a statistic describing effect strength (for example, accuracy resulting from a searchlight analysis). The code reads the 3d NIfTI images into a 4d array (people in the fourth dimension), then performs a t-test at each voxel, saving the t-values for each voxel in a new NIfTI image. This figure shows the t-value image produced by the demo code.
The demo code linked in this post does a voxelwise t-test in R. It takes as input a set of 3d NIfTI files, where each NIfTI is assumed to come from a different person, each voxel of which contains a statistic describing effect strength (for example, accuracy resulting from a searchlight analysis). The code reads the 3d NIfTI images into a 4d array (people in the fourth dimension), then performs a t-test at each voxel, saving the t-values for each voxel in a new NIfTI image. This figure shows the t-value image produced by the demo code.
 The demo code linked in this post does a voxelwise t-test in R. It takes as input a set of 3d NIfTI files, where each NIfTI is assumed to come from a different person, each voxel of which contains a statistic describing effect strength (for example, accuracy resulting from a searchlight analysis). The code reads the 3d NIfTI images into a 4d array (people in the fourth dimension), then performs a t-test at each voxel, saving the t-values for each voxel in a new NIfTI image. This figure shows the t-value image produced by the demo code.
The demo code linked in this post does a voxelwise t-test in R. It takes as input a set of 3d NIfTI files, where each NIfTI is assumed to come from a different person, each voxel of which contains a statistic describing effect strength (for example, accuracy resulting from a searchlight analysis). The code reads the 3d NIfTI images into a 4d array (people in the fourth dimension), then performs a t-test at each voxel, saving the t-values for each voxel in a new NIfTI image. This figure shows the t-value image produced by the demo code.UPDATE 4 June 2021:
The original version of this post continues below. I wrote a new version of the code to 1) avoid aaply (memory usage a bit better), and 2) use the RNifti instead of oro.nifti library (RNifti functions can be dramatically faster, and have some nice features; I now use RNifti exclusively for nifti image R input/output). I also added the files to the MVPA Meanderings OSF site, under the "voxelwise" subdirectory (direct link to R script).
Both versions of this code loop through all of the image voxels, which is most assuredly not the most efficient (or foolproof) way to perform a voxelwise t-test (I would recommend 3dttest++ instead, perhaps following 3dbucket; calling afni functions directly from R can make things quite smooth for those of us that are not bash fluent). I rewrote rather than removed this demo, however, because sometimes looping through all the voxels like this is the most straightforward way to get something done.
Monday, September 15, 2014
Connectome Workbench: 1st Steps
Here's my advice for getting started with the Connectome Workbench:
- First, go through my tutorial on getting started with Workbench: it describes downloading Workbench, starting it, loading images, and viewing overlay and underlay images.
- This tutorial introduces wb_command, and shows how to use it to create a surface file from a volumetric NIfTI (for quick-viewing purposes, not for further analysis).
- Read my summary of the different file types.
- Try the official Workbench tutorial (or at least look through the manual to get an idea of the possibilities).
- Look at my post on using the Workbench with volumetric images.
This screenshot shows scenes in action: clicking the little button marked with yellow arrows brings up the scene dialog box. I have three scenes stored in this file, and selecting one for display changes Workbench to recreate exactly how it was when the scene was created: window size, colors and scaling, loaded images, tab layout. Creating scenes for each image that might be used in a publication can save massive amounts of time: need to adjust a threshold or change a color? Just bring up the scene and make the change, no need to start from the beginning.
UPDATE 26 March 2020: linked to the new getting-started tutorial.
UPDATE 8 February 2018: linked to the new volume to surface tutorial; fixed the official tutorial link.
UPDATE 2 August 2017: removed the (very outdated!) mention of the release of Connectome Workbench 1.0; renamed this post to the more general "Connectome Workbench: 1st Steps".
Monday, September 8, 2014
nice methods: Manelis and Reder 2013, "He Who is Well Prepared ..."
It's always great to read a paper with interesting methodology clearly explained, and Manelis and Reder 2013, "He Who Is Well Prepared Has Half Won The Battle: An fMRI Study of Task Preparation" is one of those papers (full citation below). As usual, I'm not going to fully describe the paper (go read it!), but just comment on a few things that caught my eye.
First, I was struck again by the strength and consistency of the activations and deactivations associated with the n-back task; they seem as reliable as those from some motor and somatosensory tasks. The authors used a mass-univariate analysis to identify a set of ROIs to use for the MVPA, shown in this part of Figure 2 (warm colors for regions that increased with n-back level, and cool colors for regions that decreased). As the authors properly point out, doing MVPA on the task blocks with these ROIs would be somewhat circular (since a mass-univariate analysis of the task blocks was used to create the ROIs), but their main MVPA avoids circularity, since it was done on a different part of the task.
Next, I appreciated the discussions of possible confounds in the results section: the authors report pairwise accuracies, not just the three-way, explaining that they want to make sure one very accurate pair is not driving the results, and they performed a nice control analysis using randomly-selected rest volumes.
Finally, they found a correlation between classification accuracy (MVPA during task preparation periods) and behavioral performance (participant speed on the n-back task); there are still relatively few reports tying fMRI analyses to behavior, and it's nice to see another one.
 Manelis, A., & Reder, L. (2013). He Who Is Well Prepared Has Half Won The Battle: An fMRI Study of Task Preparation Cerebral Cortex DOI: 10.1093/cercor/bht262
Manelis, A., & Reder, L. (2013). He Who Is Well Prepared Has Half Won The Battle: An fMRI Study of Task Preparation Cerebral Cortex DOI: 10.1093/cercor/bht262
First, I was struck again by the strength and consistency of the activations and deactivations associated with the n-back task; they seem as reliable as those from some motor and somatosensory tasks. The authors used a mass-univariate analysis to identify a set of ROIs to use for the MVPA, shown in this part of Figure 2 (warm colors for regions that increased with n-back level, and cool colors for regions that decreased). As the authors properly point out, doing MVPA on the task blocks with these ROIs would be somewhat circular (since a mass-univariate analysis of the task blocks was used to create the ROIs), but their main MVPA avoids circularity, since it was done on a different part of the task.
Next, I appreciated the discussions of possible confounds in the results section: the authors report pairwise accuracies, not just the three-way, explaining that they want to make sure one very accurate pair is not driving the results, and they performed a nice control analysis using randomly-selected rest volumes.
Finally, they found a correlation between classification accuracy (MVPA during task preparation periods) and behavioral performance (participant speed on the n-back task); there are still relatively few reports tying fMRI analyses to behavior, and it's nice to see another one.
Subscribe to:
Posts (Atom)







